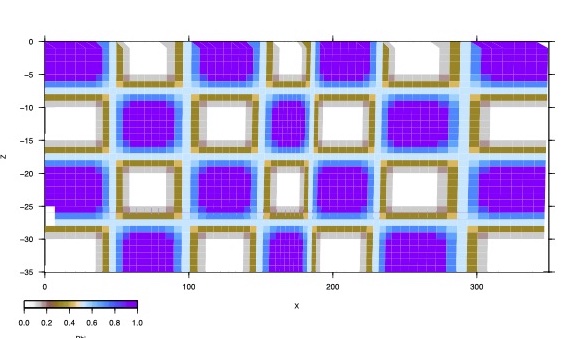
CHK1: Locking pattern for forward model
Steps Set up your faults (FA:), blocks (BL:), poles (PO:), etc. as usual. Add data using GP: or IS: Poles should be non-zero (except reference) and close to your final estimates Assign model name chk1 #checkerboard forward model 1 mo: chk1 #For example, the fault (for this example fault 9) has 14 nodes along strike and 8 downdip. # Set up the forward calculation by assigning locking indices in a checkerboard pattern. This # example uses 4 nodes (2x2) for each patch of uniform locking ft: 9 0 0 0 0 nng: 9 14 8 1 1 2 2 1 1 2 2 1 1 2 2 1 1 1 1 2 2 1 1 2 2 1 1 2 2 1 1 2 2 1 1 2 2 1 1 2 2 1 1 2 2 2 2 1 1 2 2 1 1 2 2 1 1 2 2 1 1 2 2 1 1 2 2 1 1 2 2 1 1 1 1 2 2 1 1 2 2 1 1 2 2 1 1 2 2 1 1 2 2 1 1 2 2 1 1 2 2 2 2 1 1 2 2 1 1 2 2 1 1 2 2 #Assign full locking to nodes with index 1 and no locking to nodes with index 2. nv: 9 1.0 0.0 #Set flags so fault 9 has locking but is not inverted. ff: 9 fi: -9 #Set iteration control for forward model only. ic: 0 #Don't adjust poles pi: 0 #The forward runs will produce estimated values of all your data. You add random #uncertainties to the estimates using the +rnd flag: fl: +rnd #This produces files in the chk1 directory ending with _rand.vec and _rand.insar #You can reverse the locking pattern easily by: nv: 9 0.0 1.0

CHK1: Locking pattern for forward model
#Solving for locking using randomized data # chk2 inverse model for checkerboard chk1, InSAR plus GPS mo: chk2 pf: "chk2/pio" 3 fi: 9 ff: 9 ft: 9 0 0 0 0 # Assign nodes in checkerboard pattern, but each 2x2 set of nodes has # different index nng: 9 14 8 1 1 2 2 3 3 4 4 5 5 6 6 7 7 1 1 2 2 3 3 4 4 5 5 6 6 7 7 8 8 9 9 10 10 11 11 12 12 13 13 14 14 8 8 9 9 10 10 11 11 12 12 13 13 14 14 15 15 16 16 17 17 18 18 19 19 20 20 21 21 15 15 16 16 17 17 18 18 19 19 20 20 21 21 22 22 23 23 24 24 25 25 26 26 27 27 28 28 22 22 23 23 24 24 25 25 26 26 27 27 28 28 #Starting locking model values nvg: 9 14 8 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 # remove data already read in cl: gps ins # poles to estimate (optional) pi: 2 # read randomized gps vectors gp1: CMNC "chk1/chk1_rand.vec" 1 1.0 0 0 0 1999.0 2016.0 0.3 2.5 0 1 1 0 # read randomized InSAR (INS1 is the name assigned to InSAR data in IS: for forward run) is4: DESM "chk1/chk1_INS1_rand.insar" 1 1.0 2016 2017 0 0 3 0 0 0 2.0 #iteration control ic: 1 2 1 2 em:
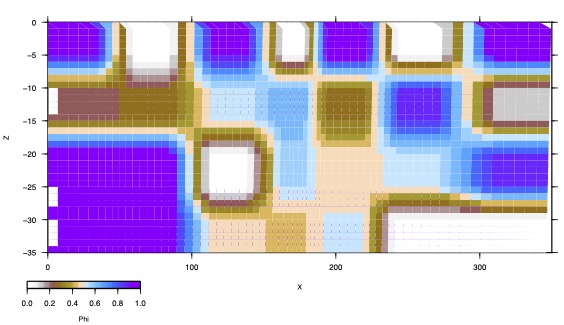
CHK2: Recovered Locking pattern for model chk1